-
Academics
-
Admissions
 Your journey to NYUAD starts here. Attend an application workshop or information session.Admissions Events
Your journey to NYUAD starts here. Attend an application workshop or information session.Admissions Events -
Research
-
Campus Life
 Live the possibilities. Be part of a dynamic community of students from over 115 countries.Take a Tour
Live the possibilities. Be part of a dynamic community of students from over 115 countries.Take a Tour - Public Programs
-
About
Exceptional education. World class research. Community-driven.Our Story
- News
- Events
- Social Media Directory
- Press Room
-
- Torch at NYUAD
- Faculty
- Current Students
- Alumni
- عربي
Algal Genomics and Synthetic Biology
Our projects are aimed at isolating locally adapted algal species and revealing their mechanisms for high salt, intense light, and drought tolerance. The obtained biological and informational resources are expected to facilitate the development of biotechnological strategies for locally and regionally relevant blue economy and food security goals. We are currently concentrating our efforts toward achieving these goals through projects outlined below:
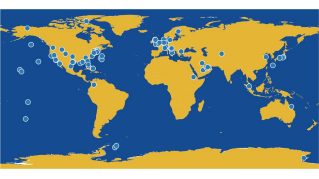
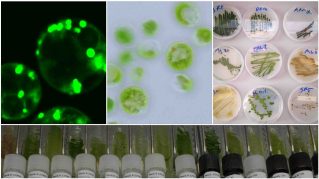
AlgallCODE: We are carrying out large-scale whole-genome sequencing of locally isolated and selected micro- and macroalgae to define their genomically encoded DNA elements (algallCODE: algae, all, Coded DNA Elements). In algallCODEmicro, we have sequenced 22 subtropical microalgal species that we isolated from the UAE and sequenced 107 additional species obtained from international algal culture collections, including UTEX (www.utex.org) and the Bigelow National Center for Algae and Marine Microbiota (https://ncma.bigelow.org/). We have analyzed these genomes comparatively, defining genomic signatures for desert acclimatization, salt tolerance, as well as the embedded records of past viral infections.
Our microalgal genomic project is complemented by exploring local and non-local macroalgae: AlgallCODEmacro is an international collaborative project addressing the lack of available macroalgal genome sequences as there are only ten genomes publicly accessible presently. We are aiming to expand the available sequences by 20 fold, with equal representation from green, brown, and red macroalgal species. We plan to deliver comparative genomics of ~200 genomes, inclusive of 30~50 local species that we are isolating from the UAE, by the end of 2022. The obtained information will enable us to identify unique adaptive patterns that influence their distribution and tolerance to extreme environments ranging from the Arabian Gulf to Polar regions. The nutritional contents of our local isolates will be determined to assess their potential use in aquaculture and as animal feed, in line with local and regional food security initiatives.
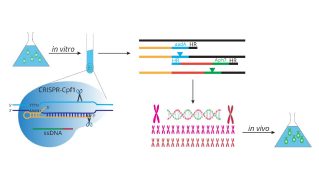
AlgalReCODE: Recent advances in DNA synthesis technology and genome editing are enabling the re-writing and re-coding of genomes, adding new dimensions to biological and biotechnological research. However, these technologies have not been implemented in any micro- or macroalgal species. In algalReCODEmicro, we are developing synthetic genomic technologies for green unicellular microalgae to understand the biology of their genomes better. We have set up pilot studies to introduce small segments of synthetic DNA into the genome of Chlamydomonas reinhardtii, a model green microalgal species. We plan to continue these pilots to replace large segments of the genome sequentially and introduce new design features in the genome.
Synthetic interactome mapping of phytoplankton: The goal of this study is to carry out a systems-level analysis of two marine cyanobacteria, Prochlorococcus marinus MED4 and P. marinus NATL1A, to define their adaptive strategies to high and low light conditions. We have completed pilot studies to map the interactions of proteins encoded in the genomes of these two species and synthetically generated and cloned nearly all their protein-coding genes, producing ~4000 clones. We are now carrying out ultra-high-throughput en mase interactome mapping of proteins encoded in P. marinus MED4 genome and will continue our work on the NATL1A strain. The resulting network maps will define convergence and rewiring of the interactions in the two species, defining their adaptive strategies to life under low and high intensities of light.
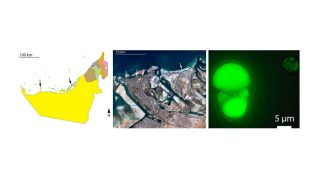
Optimization of palmitic acid production in algae: The production of palm oil in response to the global demands for this valuable oil has been often associated with environmentally damaging farming practices such as the burning of tropical forests to clear land. The ensuing loss of biodiversity, the increase in air pollution, and soil damage has impacted human and planetary health. This project aims at enhancing the production of a palm oil alternative in a locally isolated alga, Chloroidium sp. UTEX 3007, that naturally produces lipids similar to palm oil. The Chloroidium is well adapted to the UAE environment, as evidenced by its repeated isolation from different UAE locations by our group. The alga can grow in high salinity conditions, making it suitable for large-scale cultivation in the UAE or regions with a similar climate. To enhance oil production in this alga, we are using a workflow of iterative UV mutagenesis and sorting of high-lipid producing mutants by fluorescence-activated cell sorting (FACS) to select for high-lipid producing mutants. Once a superior oil-producing variant is obtained, the strain can be tested for large-scale cultivation. Because we are using a natural mutagenesis process, i.e., UV light, and no foreign DNA is introduced in the alga, the optimized strain will not be considered a GMO.
Learn more about Kourosh Salehi-Ashtiani's Laboratory of Algal Systems and Synthetic Biology.